publications
2025
- Development and validation of an autonomous artificial intelligence agent for clinical decision-making in oncologyDyke Ferber, Omar S. M. El Nahhas, Georg Wölflein, Isabella C. Wiest, Jan Clusmann, Marie-Elisabeth Leßman, Sebastian Foersch, Jacqueline Lammert, Maximilian Tschochohei, Dirk Jäger, Manuel Salto-Tellez, Nikolaus Schultz, Daniel Truhn, and Jakob Nikolas KatherNature Cancer, Jun 2025
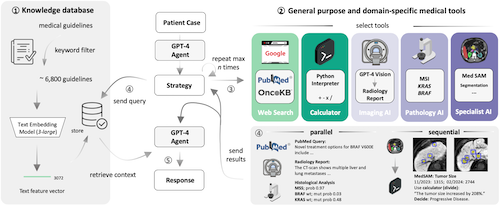
Clinical decision-making in oncology is complex, requiring the integration of multimodal data and multidomain expertise. We developed and evaluated an autonomous clinical artificial intelligence (AI) agent leveraging GPT-4 with multimodal precision oncology tools to support personalized clinical decision-making. The system incorporates vision transformers for detecting microsatellite instability and KRAS and BRAF mutations from histopathology slides, MedSAM for radiological image segmentation and web-based search tools such as OncoKB, PubMed and Google. Evaluated on 20 realistic multimodal patient cases, the AI agent autonomously used appropriate tools with 87.5% accuracy, reached correct clinical conclusions in 91.0% of cases and accurately cited relevant oncology guidelines 75.5% of the time. Compared to GPT-4 alone, the integrated AI agent drastically improved decision-making accuracy from 30.3% to 87.2%. These findings demonstrate that integrating language models with precision oncology and search tools substantially enhances clinical accuracy, establishing a robust foundation for deploying AI-driven personalized oncology support systems.
- Unsupervised Foundation Model-Agnostic Slide-Level Representation LearningTim Lenz, Peter Neidlinger, Marta Ligero, Georg Wölflein, Marko Treeck, and Jakob Nikolas KatherIn IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR), Jun 2025accepted
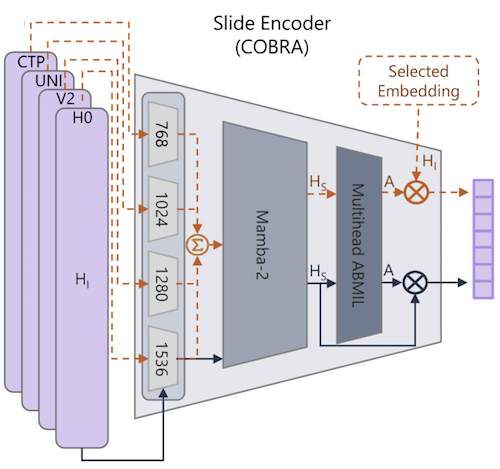
Representation learning of pathology whole-slide images (WSIs) has primarily relied on weak supervision with Multiple Instance Learning (MIL). This approach leads to slide representations highly tailored to a specific clinical task. Self-supervised learning (SSL) has been successfully applied to train histopathology foundation models (FMs) for patch embedding generation. However, generating patient or slide level embeddings remains challenging. Existing approaches for slide representation learning extend the principles of SSL from patch level learning to entire slides by aligning different augmentations of the slide or by utilizing multimodal data. By integrating tile embeddings from multiple FMs, we propose a new single modality SSL method in feature space that generates useful slide representations. Our contrastive pretraining strategy, called COBRA, employs multiple FMs and an architecture based on Mamba-2. COBRA exceeds performance of state-of-the-art slide encoders on four different public CPTAC cohorts on average by at least +3.8% AUC, despite only being pretrained on 3048 WSIs from TCGA. Additionally, COBRA is readily compatible at inference time with previously unseen feature extractors.
- Abnormality-Driven Representation Learning for Radiology ImagingMarta Ligero, Tim Lenz, Georg Wölflein, Omar S. M. El Nahhas, Daniel Truhn, and Jakob Nikolas KatherIn International Conference on Medical Image Computing and Computer-Assisted Intervention (MICCAI), Sep 2025accepted
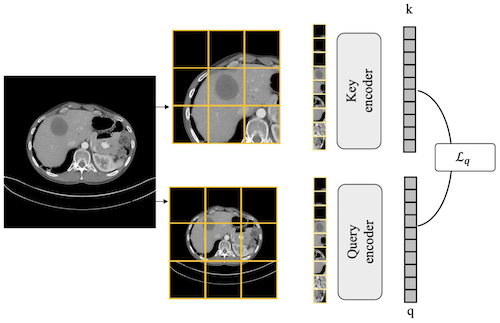
To date, the most common approach for radiology deep learning pipelines is the use of end-to-end 3D networks based on models pre-trained on other tasks, followed by fine-tuning on the task at hand. In contrast, adjacent medical fields such as pathology, which focus on 2D images, have effectively adopted task-agnostic foundational models based on self-supervised learning (SSL), combined with weakly-supervised deep learning (DL). However, the field of radiology still lacks task-agnostic representation models due to the computational and data demands of 3D imaging and the anatomical complexity inherent to radiology scans. To address this gap, we propose CLEAR, a framework for radiology images that uses extracted embeddings from 2D slices along with attention-based aggregation for efficiently predicting clinical endpoints. As part of this framework, we introduce lesion-enhanced contrastive learning (LeCL), a novel approach to obtain visual representations driven by abnormalities in 2D axial slices across different locations of the CT scans. Specifically, we trained single-domain contrastive learning approaches using three different architectures: Vision Transformers, Vision State Space Models and Gated Convolutional Neural Networks. We evaluate our approach across three clinical tasks: tumor lesion location, lung disease detection, and patient staging, benchmarking against four state-of-the-art foundation models, including BiomedCLIP. Our findings demonstrate that CLEAR using representations learned through LeCL, outperforms existing foundation models, while being substantially more compute- and data-efficient.
- LLM Agents Making Agent ToolsGeorg Wölflein, Dyke Ferber, Daniel Truhn, Ognjen Arandjelović, and Jakob Nikolas KatherIn Proceedings of the Annual Meeting of the Association for Computational Linguistics (ACL), Jul 2025accepted
Tool use has turned large language models (LLMs) into powerful agents that can perform complex multi-step tasks by dynamically utilising external software components. However, these tools must be implemented in advance by human developers, hindering the applicability of LLM agents in domains which demand large numbers of highly specialised tools, like in life sciences and medicine. Motivated by the growing trend of scientific studies accompanied by public code repositories, we propose ToolMaker, a novel agentic framework that autonomously transforms papers with code into LLM-compatible tools. Given a short task description and a repository URL, ToolMaker autonomously installs required dependencies and generates code to perform the task, using a closed-loop self-correction mechanism to iteratively diagnose and rectify errors. To evaluate our approach, we introduce a benchmark comprising 15 diverse and complex computational tasks spanning both medical and non-medical domains with over 100 unit tests to objectively assess tool correctness and robustness. ToolMaker correctly implements 80% of the tasks, substantially outperforming current state-of-the-art software engineering agents. ToolMaker therefore is a step towards fully autonomous agent-based scientific workflows.
2024
- GPT-4 for Information Retrieval and Comparison of Medical Oncology GuidelinesDyke Ferber, Isabella C. Wiest, Georg Wölflein, Matthias P. Ebert, Gernot Beutel, Jan-Niklas Eckardt, Daniel Truhn, Christoph Springfeld, Dirk Jäger, and Jakob Nikolas KatherNEJM AI, May 2024
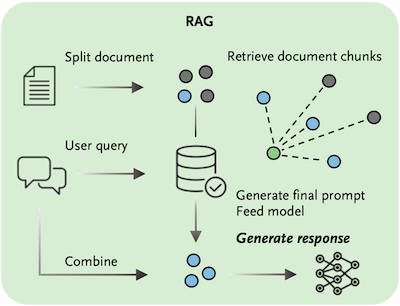
Oncologists face increasingly complex clinical decision-making processes as new cancer therapies are approved and treatment guidelines are revised at an unprecedented rate. With the aim of improving oncologists’ efficiency and supporting their adherence to the most recent treatment recommendations, we evaluated the use of the large language model generative pretrained transformer 4 (GPT-4) to interpret guidelines from the American Society of Clinical Oncology and the European Society for Medical Oncology. The ability of GPT-4 to answer clinically relevant questions regarding the management of patients with pancreatic cancer, metastatic colorectal cancer, and hepatocellular carcinoma was assessed. We also assessed GPT-4 outputs with and without retrieval-augmented generation (RAG), which provided additional knowledge to the model, and then manually compared the results with the original guideline documents. GPT-4 with RAG provided correct responses in 84% of cases (of 218 statements, 184 were correct, 30 were inaccurate, and 4 were wrong). GPT-4 without RAG provided correct responses in only 57% of cases (of 163 statements, 93 were correct, 29 were inaccurate, and 41 were wrong). We showed that GPT-4, when enhanced with additional clinical information through RAG, can accurately identify detailed similarities and disparities in diagnostic and treatment proposals across different authoritative sources. Generative pretrained transformer 4, together with retrieval-augmented generation, can precisely extract and compare medical guideline recommendations in oncology from different associations.
- From Whole-slide Image to Biomarker Prediction: End-to-End Weakly Supervised Deep Learning in Computational PathologyOmar S. M. El Nahhas, Marko Treeck, Georg Wölflein, Michaela Unger, Marta Ligero, Tim Lenz, Sophia J. Wagner, Katherine J. Hewitt, Firas Khader, Sebastian Foersch, Daniel Truhn, and Jakob Nikolas KatherNature Protocols, Sep 2024
Hematoxylin- and eosin-stained whole-slide images (WSIs) are the foundation of diagnosis of cancer. In recent years, development of deep learning-based methods in computational pathology has enabled the prediction of biomarkers directly from WSIs. However, accurately linking tissue phenotype to biomarkers at scale remains a crucial challenge for democratizing complex biomarkers in precision oncology. This protocol describes a practical workflow for solid tumor associative modeling in pathology (STAMP), enabling prediction of biomarkers directly from WSIs by using deep learning. The STAMP workflow is biomarker agnostic and allows for genetic and clinicopathologic tabular data to be included as an additional input, together with histopathology images. The protocol consists of five main stages that have been successfully applied to various research problems: formal problem definition, data preprocessing, modeling, evaluation and clinical translation. The STAMP workflow differentiates itself through its focus on serving as a collaborative framework that can be used by clinicians and engineers alike for setting up research projects in the field of computational pathology. As an example task, we applied STAMP to the prediction of microsatellite instability (MSI) status in colorectal cancer, showing accurate performance for the identification of tumors high in MSI. Moreover, we provide an open-source code base, which has been deployed at several hospitals across the globe to set up computational pathology workflows. The STAMP workflow requires one workday of hands-on computational execution and basic command line knowledge.
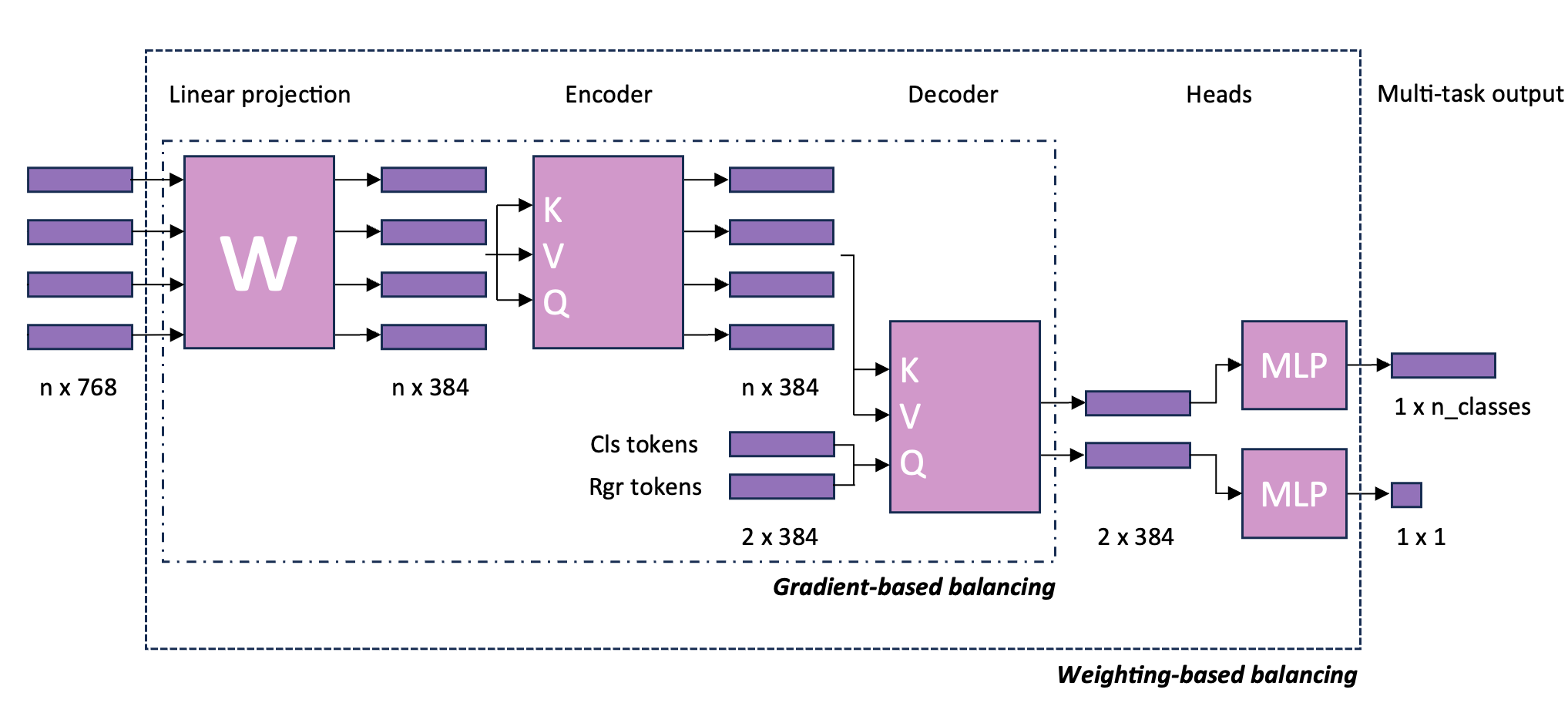
- Joint multi-task learning improves weakly-supervised biomarker prediction in computational pathologyOmar S. M. El Nahhas, Georg Wölflein, Marta Ligero, Tim Lenz, Marko Treeck, Firas Khader, Daniel Truhn, and Jakob Nikolas KatherIn International Conference on Medical Image Computing and Computer-Assisted Intervention (MICCAI), Oct 2024
Deep Learning (DL) can predict biomarkers directly from digitized cancer histology in a weakly-supervised setting. Recently, the prediction of continuous biomarkers through regression-based DL has seen an increasing interest. Nonetheless, clinical decision making often requires a categorical outcome. Consequently, we developed a weakly-supervised joint multi-task Transformer architecture which has been trained and evaluated on four public patient cohorts for the prediction of two key predictive biomarkers, microsatellite instability (MSI) and homologous recombination deficiency (HRD), trained with auxiliary regression tasks related to the tumor microenvironment. Moreover, we perform a comprehensive benchmark of 16 approaches of task balancing for weakly-supervised joint multi-task learning in computational pathology. Using our novel approach, we improve over the state-of-the-art area under the receiver operating characteristic by +7.7% and +4.1%, as well as yielding better clustering of latent embeddings by +8% and +5% for the prediction of MSI and HRD in external cohorts, respectively.
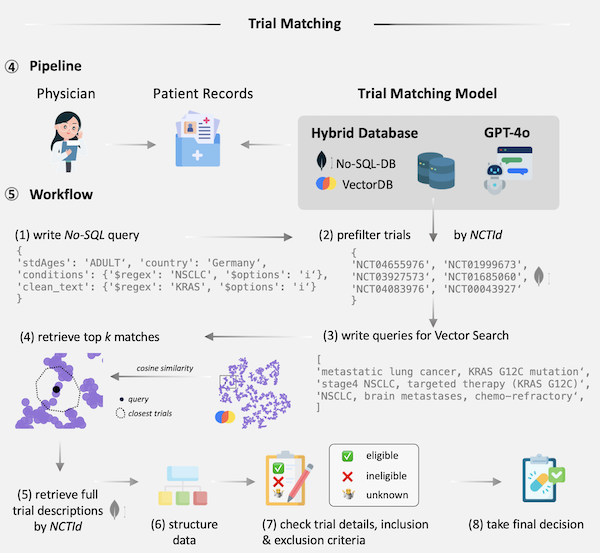
- End-To-End Clinical Trial Matching with Large Language ModelsDyke Ferber, Lars Hilgers, Isabella C Wiest, Marie-Elisabeth Leßmann, Jan Clusmann, Peter Neidlinger, Jiefu Zhu, Georg Wölflein, Jacqueline Lammert, Maximilian Tschochohei, Heiko Böhme, Dirk Jäger, Mihaela Aldea, Daniel Truhn, Christiane Höper, and Jakob Nikolas KatherJul 2024under review
Matching cancer patients to clinical trials is essential for advancing treatment and patient care. However, the inconsistent format of medical free text documents and complex trial eligibility criteria make this process extremely challenging and time-consuming for physicians. We investigated whether the entire trial matching process – from identifying relevant trials among 105,600 oncology-related clinical trials on clinicaltrials.gov to generating criterion-level eligibility matches – could be automated using Large Language Models (LLMs). Using GPT-4o and a set of 51 synthetic Electronic Health Records (EHRs), we demonstrate that our approach identifies relevant candidate trials in 93.3% of cases and achieves a preliminary accuracy of 88.0% when matching patient-level information at the criterion level against a baseline defined by human experts. Utilizing LLM feedback reveals that 39.3% criteria that were initially considered incorrect are either ambiguous or inaccurately annotated, leading to a total model accuracy of 92.7% after refining our human baseline. In summary, we present an end-to-end pipeline for clinical trial matching using LLMs, demonstrating high precision in screening and matching trials to individual patients, even outperforming the performance of qualified medical doctors. Our fully end-to-end pipeline can operate autonomously or with human supervision and is not restricted to oncology, offering a scalable solution for enhancing patient-trial matching in real-world settings.
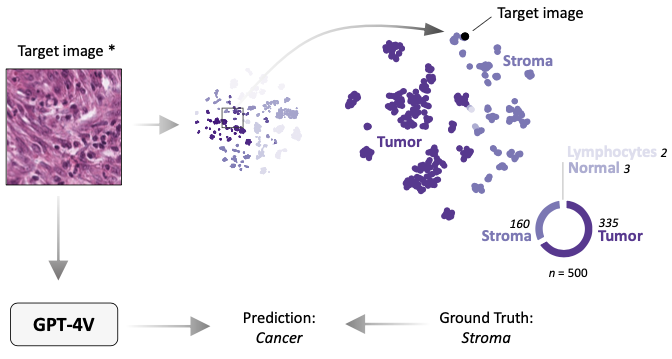
- In-context learning enables multimodal large language models to classify cancer pathology imagesDyke Ferber, Georg Wölflein, Isabella C. Wiest, Marta Ligero, Srividhya Sainath, Narmin Ghaffari Laleh, Omar S. M. El Nahhas, Gustav Müller-Franzes, Dirk Jäger, Daniel Truhn, and Jakob Nikolas KatherNature Communications, Nov 2024
Medical image classification requires labeled, task-specific datasets which are used to train deep learning networks de novo, or to fine-tune foundation models. However, this process is computationally and technically demanding. In language processing, in-context learning provides an alternative, where models learn from within prompts, bypassing the need for parameter updates. Yet, in-context learning remains underexplored in medical image analysis. Here, we systematically evaluate the model Generative Pretrained Transformer 4 with Vision capabilities (GPT-4V) on cancer image processing with in-context learning on three cancer histopathology tasks of high importance: Classification of tissue subtypes in colorectal cancer, colon polyp subtyping and breast tumor detection in lymph node sections. Our results show that in-context learning is sufficient to match or even outperform specialized neural networks trained for particular tasks, while only requiring a minimal number of samples. In summary, this study demonstrates that large vision language models trained on non-domain specific data can be applied out-of-the box to solve medical image-processing tasks in histopathology. This democratizes access of generalist AI models to medical experts without technical background especially for areas where annotated data is scarce.
- A Good Feature Extractor Is All You Need for Weakly Supervised Pathology Slide ClassificationGeorg Wölflein, Dyke Ferber, Asier Rabasco Meneghetti, Omar S. M. El Nahhas, Daniel Truhn, Zunamys I. Carrero, David J. Harrison, Ognjen Arandjelović, and Jakob Nikolas KatherIn European Conference on Computer Vision (ECCV), Sep 2024BioImage Computing Workshop (oral)
Stain normalisation is thought to be a crucial preprocessing step in computational pathology pipelines. We question this belief in the context of weakly supervised whole slide image classification, motivated by the emergence of powerful feature extractors trained using self-supervised learning on diverse pathology datasets. To this end, we performed the most comprehensive evaluation of publicly available pathology feature extractors to date, involving more than 8,000 training runs across nine tasks, five datasets, three downstream architectures, and various preprocessing setups. Notably, we find that omitting stain normalisation and image augmentations does not compromise downstream slide-level classification performance, while incurring substantial savings in memory and compute. Using a new evaluation metric that facilitates relative downstream performance comparison, we identify the best publicly available extractors, and show that their latent spaces are remarkably robust to variations in stain and augmentations like rotation. Contrary to previous patch-level benchmarking studies, our approach emphasises clinical relevance by focusing on slide-level biomarker prediction tasks in a weakly supervised setting with external validation cohorts. Our findings stand to streamline digital pathology workflows by minimising preprocessing needs and informing the selection of feature extractors.
- Benchmarking Pathology Feature Extractors for Whole Slide Image ClassificationGeorg Wölflein, Dyke Ferber, Asier Rabasco Meneghetti, Omar S. M. El Nahhas, Daniel Truhn, Zunamys I. Carrero, David J. Harrison, Ognjen Arandjelović, and Jakob Nikolas KatherJun 2024
Weakly supervised whole slide image classification is a key task in computational pathology, which involves predicting a slide-level label from a set of image patches constituting the slide. Constructing models to solve this task involves multiple design choices, often made without robust empirical or conclusive theoretical justification. To address this, we conduct a comprehensive benchmarking of feature extractors to answer three critical questions: 1) Is stain normalisation still a necessary preprocessing step? 2) Which feature extractors are best for downstream slide-level classification? 3) How does magnification affect downstream performance? Our study constitutes the most comprehensive evaluation of publicly available pathology feature extractors to date, involving more than 10,000 training runs across 14 feature extractors, 9 tasks, 5 datasets, 3 downstream architectures, 2 levels of magnification, and various preprocessing setups. Our findings challenge existing assumptions: 1) We observe empirically, and by analysing the latent space, that skipping stain normalisation and image augmentations does not degrade performance, while significantly reducing memory and computational demands. 2) We develop a novel evaluation metric to compare relative downstream performance, and show that the choice of feature extractor is the most consequential factor for downstream performance. 3) We find that lower-magnification slides are sufficient for accurate slide-level classification. Contrary to previous patch-level benchmarking studies, our approach emphasises clinical relevance by focusing on slide-level biomarker prediction tasks in a weakly supervised setting with external validation cohorts. Our findings stand to streamline digital pathology workflows by minimising preprocessing needs and informing the selection of feature extractors.
2023
- Performance comparison between federated and centralized learning with a deep learning model on Hoechst stained imagesDamien Alouges, Georg Wölflein, In Hwa Um, David Harrison, Ognjen Arandjelović, Christophe Battail, and Stéphane GazutJul 2023
Medical data is not fully exploited by Machine Learning (ML) techniques because the privacy concerns restrict the sharing of sensitive information and consequently the use of centralized ML schemes. Usually, ML models trained on local data are failing to reach their full potential owing to low statistical power. Federated Learning (FL) solves critical issues in the healthcare domain such as data privacy and enables multiple contributors to build a common and robust ML model by sharing local learning parameters without sharing data. FL approaches are mainly evaluated in the literature using benchmarks and the trade-off between accuracy and privacy still has to be more studied in a realistic clinical context. In this work, part of the European project KATY (GA:101017453), we evaluate this trade-off for a CD3/CD8 cells staining procedure from Hoechst images. Wölflein et al. developed a deep learning GAN model that synthesizes CD3 and CD8 stains from kidney cancer tissue slides, trained on 473,000 patches (256x256 pixels) from 8 whole slide images. We modified the training to simulate a FL approach by distributing the learning across 8 clients and aggregating the parameters to create the overall model. We present then the performance comparison between FL and centralized learning.
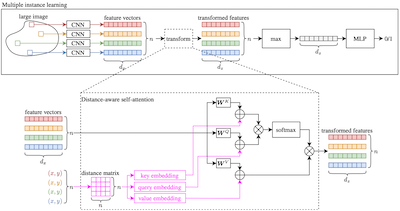
- Deep Multiple Instance Learning with Distance-Aware Self-AttentionGeorg Wölflein, Lucie Charlotte Magister, Pietro Liò, David J Harrison, and Ognjen ArandjelovićMay 2023
Traditional supervised learning tasks require a label for every instance in the training set, but in many real-world applications, labels are only available for collections (bags) of instances. This problem setting, known as multiple instance learning (MIL), is particularly relevant in the medical domain, where high-resolution images are split into smaller patches, but labels apply to the image as a whole. Recent MIL models are able to capture correspondences between patches by employing self-attention, allowing them to weigh each patch differently based on all other patches in the bag. However, these approaches still do not consider the relative spatial relationships between patches within the larger image, which is especially important in computational pathology. To this end, we introduce a novel MIL model with distance-aware self-attention (DAS-MIL), which explicitly takes into account relative spatial information when modelling the interactions between patches. Unlike existing relative position representations for self-attention which are discrete, our approach introduces continuous distance-dependent terms into the computation of the attention weights, and is the first to apply relative position representations in the context of MIL. We evaluate our model on a custom MNIST-based MIL dataset that requires the consideration of relative spatial information, as well as on CAMELYON16, a publicly available cancer metastasis detection dataset, where we achieve a test AUROC score of 0.91. On both datasets, our model outperforms existing MIL approaches that employ absolute positional encodings, as well as existing relative position representation schemes applied to MIL. Our code is available at https://anonymous.4open.science/r/das-mil.
- HoechstGAN: Virtual Lymphocyte Staining Using Generative Adversarial NetworksGeorg Wölflein, In Hwa Um, David J Harrison, and Ognjen ArandjelovićIn Proceedings of the IEEE/CVF Winter Conference on Applications of Computer Vision (WACV), Jan 2023
The presence and density of specific types of immune cells are important to understand a patient’s immune response to cancer. However, immunofluorescence staining required to identify T cell subtypes is expensive, timeconsuming, and rarely performed in clinical settings. We present a framework to virtually stain Hoechst images (which are cheap and widespread) with both CD3 and CD8 to identify T cell subtypes in clear cell renal cell carcinoma using generative adversarial networks. Our proposed method jointly learns both staining tasks, incentivising the network to incorporate mutually beneficial information from each task. We devise a novel metric to quantify the virtual staining quality, and use it to evaluate our method.
- Whole-Slide Images and Patches of Clear Cell Renal Cell Carcinoma Tissue Sections Counterstained with Hoechst 33342, CD3, and CD8 Using Multiple ImmunofluorescenceGeorg Wölflein, In Hwa Um, David J Harrison, and Ognjen ArandjelovićData, Feb 2023
In recent years, there has been an increased effort to digitise whole-slide images of cancer tissue. This effort has opened up a range of new avenues for the application of deep learning in oncology. One such avenue is virtual staining, where a deep learning model is tasked with reproducing the appearance of stained tissue sections, conditioned on a different, often times less expensive, input stain. However, data to train such models in a supervised manner where the input and output stains are aligned on the same tissue sections are scarce. In this work, we introduce a dataset of ten whole-slide images of clear cell renal cell carcinoma tissue sections counterstained with Hoechst 33342, CD3, and CD8 using multiple immunofluorescence. We also provide a set of over 600,000 patches of size 256 × 256 pixels extracted from these images together with cell segmentation masks in a format amenable to training deep learning models. It is our hope that this dataset will be used to further the development of deep learning methods for digital pathology by serving as a dataset for comparing and benchmarking virtual staining models.
2022
- Use of High-Plex Data Reveals Novel Insights into the Tumour Microenvironment of Clear Cell Renal Cell CarcinomaRaffaele De Filippis, Georg Wölflein, In Hwa Um, Peter D Caie, Sarah Warren, Andrew White, Elizabeth Suen, Emily To, Ognjen Arandjelović, and David J HarrisonCancers, Nov 2022
Although immune checkpoint inhibitors (ICIs) have significantly improved the oncological outcomes, about one-third of patients affected by clear cell renal cell carcinoma (ccRCC) still experience recurrence. Current prognostic algorithms, such as the Leibovich score (LS), rely on morphological features manually assessed by pathologists and are therefore subject to bias. Moreover, these tools do not consider the heterogeneous molecular milieu present in the Tumour Microenvironment (TME), which may have prognostic value. We systematically developed a semi-automated method to investigate 62 markers and their combinations in 150 primary ccRCCs using Multiplex Immunofluorescence (mIF), NanoString GeoMx® Digital Spatial Profiling (DSP) and Artificial Intelligence (AI)-assisted image analysis in order to find novel prognostic signatures and investigate their spatial relationship. We found that coexpression of cancer stem cell (CSC) and epithelial-to-mesenchymal transition (EMT) markers such as OCT4 and ZEB1 are indicative of poor outcome. OCT4 and the immune markers CD8, CD34, and CD163 significantly stratified patients at intermediate LS. Furthermore, augmenting the LS with OCT4 and CD34 improved patient stratification by outcome. Our results support the hypothesis that combining molecular markers has prognostic value and can be integrated with morphological features to improve risk stratification and personalised therapy. To conclude, GeoMx® DSP and AI image analysis are complementary tools providing high multiplexing capability required to investigate the TME of ccRCC, while reducing observer bias.
2021
- Determining Chess Game State from an ImageGeorg Wölflein and Ognjen ArandjelovićJournal of Imaging, Jun 2021
Identifying the configuration of chess pieces from an image of a chessboard is a problem in computer vision that has not yet been solved accurately. However, it is important for helping amateur chess players improve their games by facilitating automatic computer analysis without the overhead of manually entering the pieces. Current approaches are limited by the lack of large datasets and are not designed to adapt to unseen chess sets. This paper puts forth a new dataset synthesised from a 3D model that is an order of magnitude larger than existing ones. Trained on this dataset, a novel end-to-end chess recognition system is presented that combines traditional computer vision techniques with deep learning. It localises the chessboard using a RANSAC-based algorithm that computes a projective transformation of the board onto a regular grid. Using two convolutional neural networks, it then predicts an occupancy mask for the squares in the warped image and finally classifies the pieces. The described system achieves an error rate of 0.23% per square on the test set, 28 times better than the current state of the art. Further, a few-shot transfer learning approach is developed that is able to adapt the inference system to a previously unseen chess set using just two photos of the starting position, obtaining a per-square accuracy of 99.83% on images of that new chess set. The code, dataset, and trained models are made available online.
datasets
2022
- Whole slide images and patches of clear cell renal cell carcinoma counterstained with multiple immunofluorescence for Hoechst, CD3, and CD8Georg Wölflein, In Hwa Um, David J Harrison, and Ognjen ArandjelovićDec 2022
We provide a dataset of 10 whole slide images of clear cell renal cell carcinoma tissue sections that were counterstained with Hoechst 33342, Cy3 and Cy5. The fluorescent images were captured using Zeiss Axio Scan Z1. We used three different fluorescent channels (Hoechst 33342, Cy3 and Cy5) simultaneously to capture individual channel images under 20x object magnification with respective exposure times of 10 ms, 20 ms and 30 ms. We also provide clinical data including age at surgery, gender, disease free months, nuclear grade, and Leibovich score. Alongside the WSIs, we provide pre-processed and normalised patches (each 256x256 pixels) from the WSIs in a format that can be readily ingested by deep learning models for their training. For each patch, we provide normalised Hoechst, CD3, and CD8 images, as well as cell masks identified using the StarDist algorithm. Each cell is annotated as CD3, CD8 (a subset of CD3), or neither.
2021
theses
2021
- Determining Chess Game State From an Image Using Machine LearningGeorg Wölflein (supervised by Ognjen Arandjelović)University of St Andrews, Jan 2021
Identifying the configuration of chess pieces from an image of a chessboard is a problem in computer vision that has not yet been solved accurately. However, it is important for helping amateur chess players improve their games by facilitating automatic computer analysis without the overhead of manually entering the pieces. Current approaches are limited by the lack of large datasets and are not designed to adapt to unseen chess sets. This project puts forth a new dataset synthesised from a 3D model that is two orders of magnitude larger than existing ones. Trained on this dataset, a novel end-to-end chess recognition system is presented that combines traditional computer vision techniques with deep learning. It localises the chessboard using a RANSAC-based algorithm that computes a projective transformation of the board onto a regular grid. Using two convolutional neural networks, it then predicts an occupancy mask for the squares in the warped image and finally classifies the pieces. The described system achieves an error rate of 0.23% per square on the test set, 28 times better than the current state of the art. Further, a one-shot transfer learning approach is developed that is able to adapt the inference system to a previously unseen chess set using just two photos of the starting position, obtaining a per-square accuracy of 99.83% on images of that new chess set. Inference takes less than half a second on a GPU and about two seconds on a CPU. The feasibility of the system is demonstrated in an interactive web app available at https://www.chesscog.com.
2020
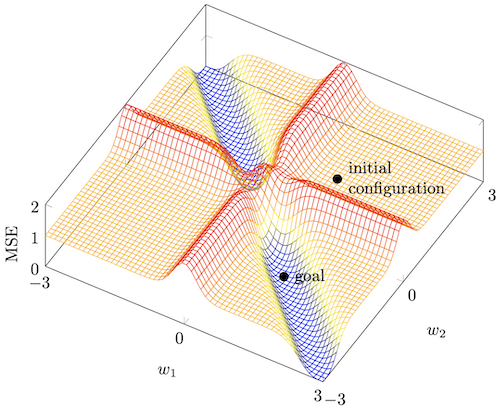
- Freeing Neural Training Through SurfingGeorg Wölflein (supervised by Michael Weir)University of St Andrews, Apr 2020
Gradient methods based on backpropagation are widely used in training multilayer feedforward neural networks. However, such algorithms often converge to suboptimal weight configurations known as local minima. This report presents a novel minimal example of the local minimum problem with only three training samples and demonstrates its suitability for investigating and resolving said problem by analysing its mathematical properties and conditions leading to the failure of conventional training regimes. A different perspective for training neural networks is introduced that concerns itself with neural spaces and is applied to study the local minimum example.This gives rise to the concept of setting intermediate subgoals during training which is demonstrated to be a viable and effective means of overcoming the local minimum problem. The versatility of subgoal-based approaches is highlighted by showing their potential for training more generally. An example of a subgoal-based training regime using sampling and an adaptive clothoid for establishing a goal-connecting path is suggested as a proof of concept for further research. In addition, this project includes the design and implementation of a software framework for monitoring the performance of different neural training algorithms on a given problem simultaneously and in real time. This framework can be used to reproduce the findings of how classical algorithms fail to find the global minimum in the aforementioned example.