Abstract
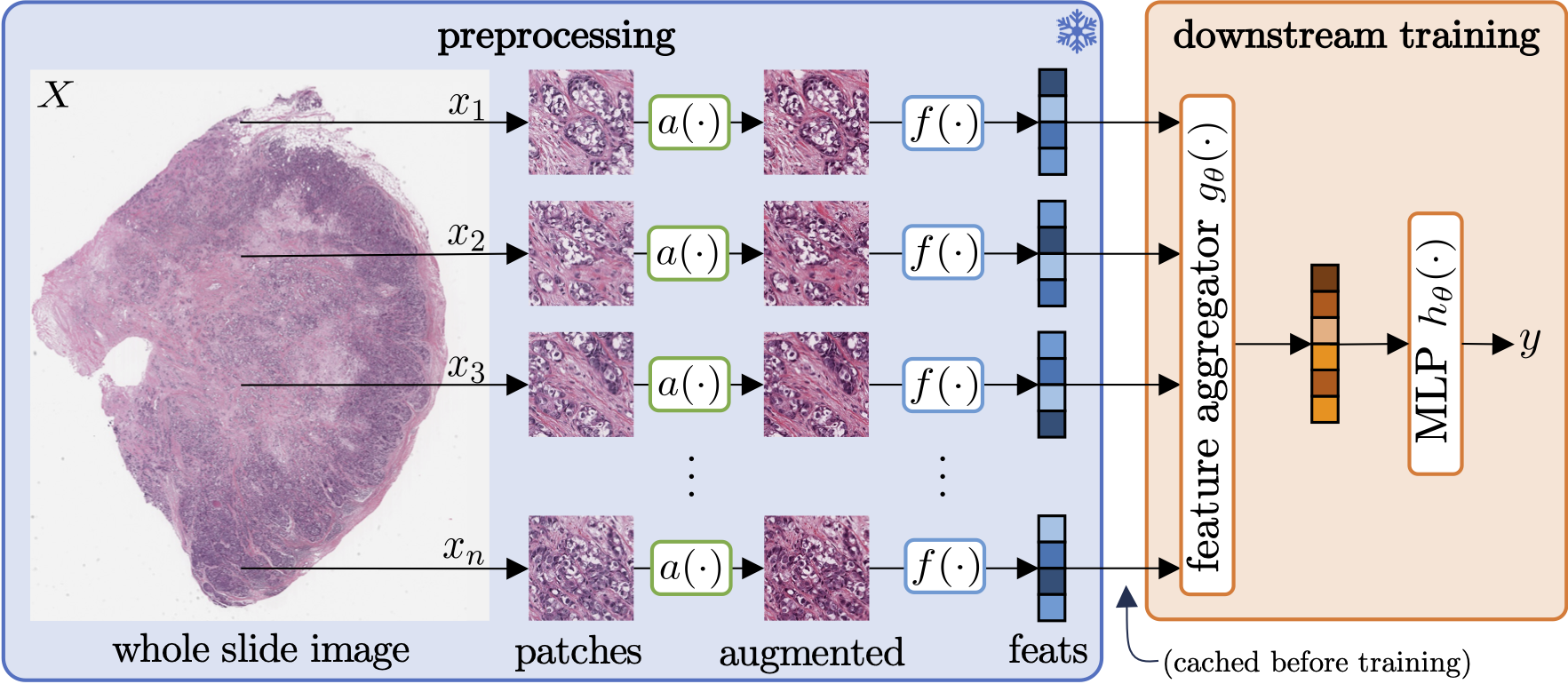
Stain normalisation is thought to be a crucial preprocessing step in computational pathology pipelines. We question this belief in the context of weakly supervised whole slide image classification, motivated by the emergence of powerful feature extractors trained using self-supervised learning on diverse pathology datasets. To this end, we performed the most comprehensive evaluation of publicly available pathology feature extractors to date, involving more than 8,000 training runs across nine tasks, five datasets, three downstream architectures, and various preprocessing setups. Notably, we find that omitting stain normalisation and image augmentations does not compromise downstream slide-level classification performance, while incurring substantial savings in memory and compute. Using a new evaluation metric that facilitates relative downstream performance comparison, we identify the best publicly available extractors, and show that their latent spaces are remarkably robust to variations in stain and augmentations like rotation. Contrary to previous patch-level benchmarking studies, our approach emphasises clinical relevance by focusing on slide-level biomarker prediction tasks in a weakly supervised setting with external validation cohorts. Our findings stand to streamline digital pathology workflows by minimising preprocessing needs and informing the selection of feature extractors.
TLDR: We perform a comprehensive empirical study on weakly supervised whole slide image classification, showing (i) stain normalisation is unnecessary no matter the choice of feature extractor, and (ii) which publicly available feature extractors are best.
Citation
If you would like to cite our work, please use:
@inproceedings{wolflein2023good,
author = {W\"{o}lflein, Georg and Ferber, Dyke and Meneghetti, Asier Rabasco and El Nahhas, Omar S. M. and Truhn, Daniel and Carrero, Zunamys I. and Harrison, David J. and Arandjelovi\'{c}, Ognjen and Kather, Jakob Nikolas},
title = {A Good Feature Extractor Is All You Need for Weakly Supervised Pathology Slide Classification},
year = {2024},
month = {09},
booktitle = {European Conference on Computer Vision (ECCV) BioImage Computing Workshop},
publisher = {Springer},
}
Acknowledgements
GW is supported by Lothian NHS.